Host-Microbe Interactions in the Human Gut
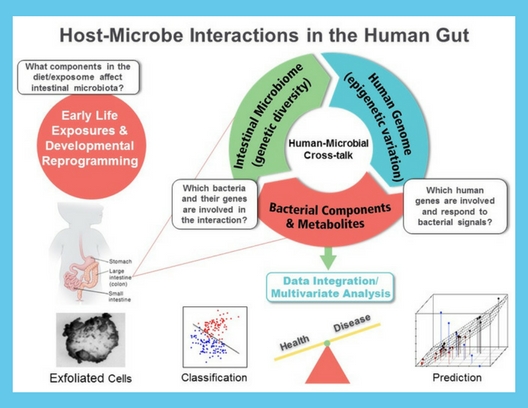
Development of Novel Exfoliated Cell Noninvasive Methodology to Monitor Host/Microbe Interactions.
Early detection can be considered a method for prevention in the sense that it can prevent serious morbidity and mortality. To address this need, our team has identified novel early detection biomarkers, e.g., genomic signals, to classify/predict chronic inflammation, metabolic profiling, immune status, and gut barrier function on a molecular level in mouse models and humans. For this purpose, we mathematically model dynamical system behavior for the purpose of deriving therapeutic strategies to alter undesirable cellular behavior. Outcomes include the prediction of new nutraceutical / drug targets based on intracellular signaling pathways. Cutting edge applications include: (i) the influences of diet and the gut microbiome on host (using exfoliated cells) epigenetic modulation in neonates, and (ii) simultaneous analysis of the multivariate structure between the gut metatranscriptome and host transcriptome using noninvasive methodologies.
- A. McLeod; P. Wolf, R.S. Chapkin, L.A. Davidson, I. Ivanov, M. Berbaum, L.R. Williams, H.R. Gaskins, J. Ridlon, J. Sanchez-Flack, L. Blumstein, L. Schiffer, A. Hamm, K. Cares, M. Antonic, B. Penalver Bernabe, M. Fitzgibbon and L. Tussing-Humphreys.Design of a six month, parallel group Mediterranean diet and weight loss randomized controlled lifestyle intervention targeting the bile acid-gut microbiome axis to reduce colorectal cancer risk among African American/Black adults with obesity. Trials 24:113, 2023. PMID: 36793105, PMCID: PMC9930092
- Personalized Nutrition Using Microbial Metabolite Phenotype to Stratify Participants and Non-Invasive Host Exfoliomics Reveal the Effects of Flaxseed Lignan Supplementation in a Placebo-Controlled Crossover Trial. Nutrients. 2022 Jun 8;14(12). doi: 10.3390/nu14122377. PubMed PMID: 35745107; PubMed Central PMCID: PMC9230005.
- Exfoliated epithelial cell transcriptome reflects both small and large intestinal cell signatures in piglets. American Journal of Physiology-Gastrointestinal and Liver Physiology 321:G41-G51, 2021. PMID:33949197, PMCID: PMC8321797
- K. He, S.M. Donovan, I. Ivanov, J. Goldsby, L.A. Davidson and R.S. Chapkin. Assessing the multivariate relationship between the human infant intestinal exfoliated cell transcriptome (exfoliome) and microbiome in response to diet. Microorganisms 8:2023, 2020. PMID:33353204, PMCID:PMC7766018
- J.W. Lampe, E. Kim, L. Levy, L.A. Davidson, J.S. Goldsby, F.L. Miles, S.L. Navarro, T.W. Randolph, N. Zhao, I. Ivanov, A.M, Kaz, C. Damman, D.M. Hockenbery, M.A.J. Hullar and R.S. Chapkin. Colonic mucosal and exfoliome transcriptomic profiling and fecal microbiome response to flaxseed lignan extract intervention in humans. American Journal of Clinical Nutrition 110:377-390, 2019. PMID:31175806, PMCID:PMC6669062
- Whitfield-Cargile, N. Cohen, K. He, I. Ivanov, J. Goldsby, A. Chamoun-Emanuelli, B. Weeks, L.A. Davidson and R.S. Chapkin. The non-invasive exfoliated transcriptome (exfoliome) reflects the tissue-level transcriptome in a mouse model of NSAID-enteropathy. Nature Scientific Reports 7:14687, 2017. PMID:29089621, PMCID:PMC5665873
- V. Seidel, M.A Azcarate-Peril, R.S. Chapkin and N.D. Turner. Shaping functional gut microbiota using dietary bioactives to reduce colon cancer risk. Seminars in Cancer Biology 46:191-204, 2017. PMID:28676459, PMCID:PMC5626600
- Y. Hou, E. Kim, Y.Y. Fan, N.R. Fuentes, L.A. Davidson and R.S. Chapkin. Nutrient-gene interaction in colon cancer, from the membrane to cellular physiology. Annual Reviews of Nutrition 36:543-570, 2016. PMID:27431370, PMCID:PMC5034935
- Wang, M. Li, C.B. Lebrilla, R.S. Chapkin, I. Ivanov and S.M. Donovan. Fecal microbiota composition of breast-fed infants differs from formula-fed and is correlated with human milk oligosaccharides consumed. Journal of Pediatric Gastroentrology & Nutrition 60:825-833, 2015. PMID:25651488 PMC4441539
- M. Knight, L.A. Davidson, D. Herman, C.R. Martin, J.S. Goldsby, I.V. Ivanov, S.M. Donovan and R.S. Chapkin. Non-invasive analysis of intestinal development in premature and full term infants using RNA-Sequencing. Scientific Reports Nature 4:5453; DOI:10.1038/srep05453, 2014. PMID:24965658 PMC4071321
- Schwartz, I. Friedberg, I.V. Ivanov, L.A. Davidson, J.S. Goldsby, D.B. Dahl, D. Herman, M. Wang, S.M. Donovan and R.S. Chapkin. A Metagenomic study of diet-dependent interaction between gut microflora and host in infants reveals differences in developmental and immune responses. Genome Biology 13:R32 doi:10.1186/gb-2012-13-4-r32, 2012. PMID:22546241 PMC3446306
- R.S. Chapkin, C. Zhao, I. Ivanov, L.A. Davidson, J.S. Goldsby, J.R. Lupton, R.A. Mathai, M. Siegel, D. Rai, M. Russell, S.M. Donovan and E.R. Dougherty. Non-invasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells. American Journal of Physiology 298:G582-G589, 2010. PMID: 20203060 PMC2867429
- C. Zhao, I. Ivanov, E.R. Dougherty, T.J. Hartman, E. Lanza, N.H. Colburn, J.R. Lupton, L.A. Davidson and R.S. Chapkin. Non-invasive detection of candidate molecular biomarkers in subjects with a history of insulin resistance and colorectal adenomas. Cancer Prevention Research 2:590-597, 2009. PMID: 19470793 PMCID:PMC2745241
